Using openFDA can be a breeze, if you know how to construct good queries. This short guide will get you started with openFDA, and show you how to put together more complicated queries. We start by loading openFDA.
#> ! OpenFDA API key set.
#> ℹ You can retrieve the key with the following code:
#> `keyring::key_get(service = "OPENFDA_KEY", user = "openFDA")`The openFDA API
The openFDA API makes public FDA data available from a simple, public API. Users of this API have access to FDA data on food, human and veterinary drugs, devices, and more. You can read all about it at their website. The API limits results sets to 1000 records. To access over 1000 results, you’d usually need to manually submit multiple requests which skip past larger and larger chunks of the dataset (see online for more info).
With this package, however, that can be handled for you. By default,
openFDA() will ask you if you want to retrieve all possible
results. However, asking for user input isn’t possible when in a
vignette. Additionally, we don’t want to do paging for these examples,
as our searches get lots of results. Therefore, to prevent paging, we
set the package-level option openFDA.paging to
"never". Now openFDA() will automatically
not perform paging, even when it’s possible.
options(openFDA.paging = "never")A simple openFDA query
The simplest way to query the openFDA API is to identify the
endpoint you want to use and provide other search
terms. For example, this snippet retrieves 2 records about
adverse events in the drugs endpoint.
The empty search string ("") means the results will be
non-specific.
search <- openFDA(search = "",
endpoint = "drug-event",
limit = 2)
#> ! openFDA returned more results (19028302) than your `limit` (2).
#> ℹ With 2 results per page, pagination will require 9,514,151 queries to
#> retrieve all of these results. This will take at least 11h 43s.
search
#> <httr2_response>
#> GET https://api.fda.gov/drug/event.json?search=&limit=2
#> Status: 200 OK
#> Content-Type: application/json
#> Body: In memory (3923 bytes)The warning given about pagination is because we searched on the
whole dataset, but used limit to only pull out the first
two results. We can suppress this warning by setting
paging_verbosity to "quiet". Now we won’t page
the whole dataset, because paging == "never", and we won’t
get that warning either:
search <- openFDA(search = "",
endpoint = "drug-event",
limit = 2,
paging = "never",
paging_verbosity = "quiet")
search
#> <httr2_response>
#> GET https://api.fda.gov/drug/event.json?search=&limit=2
#> Status: 200 OK
#> Content-Type: application/json
#> Body: In memory (3923 bytes)Instead of setting the paging and
paging_verbosity arguments each time you call the function,
it is better to set the package-level option for these behaviours. You
can do this with any function that edits options:
options(
openFDA.paging = "never",
openFDA.paging_verbosity = "quiet"
)openFDA results
The function returns an httr2 response object, with
attached JSON data. We use httr2::resp_body_json() to
extract the underlying data.
json <- httr2::resp_body_json(search)If you don’t specify a field to count on, the JSON data
has two sections - meta and results.
Meta
The meta section has important metadata on your results,
which includes:
-
disclaimer- An important disclaimer regarding the data provided by openFDA. -
license- A webpage with license terms that govern the openFDA API. -
last_updated- The last date at which this openFDA endpoint was updated. -
results.skip- How many results were skipped? Set by theskipparameter inopenFDA(). -
results.limit- How many results were retrieved? Set by thelimitparameter inopenFDA(). -
results.total- How many results were there in total matching yoursearchcriteria?
json$meta
#> $disclaimer
#> [1] "Do not rely on openFDA to make decisions regarding medical care. While we make every effort to ensure that data is accurate, you should assume all results are unvalidated. We may limit or otherwise restrict your access to the API in line with our Terms of Service."
#>
#> $terms
#> [1] "https://open.fda.gov/terms/"
#>
#> $license
#> [1] "https://open.fda.gov/license/"
#>
#> $last_updated
#> [1] "2025-04-28"
#>
#> $results
#> $results$skip
#> [1] 0
#>
#> $results$limit
#> [1] 2
#>
#> $results$total
#> [1] 19028302Results
For non-count queries, this will be a set of records
which were found in the endpoint and match your search
term.
json$results
#> [[1]]
#> [[1]]$safetyreportid
#> [1] "5801206-7"
#>
#> [[1]]$transmissiondateformat
#> [1] "102"
#>
#> [[1]]$transmissiondate
#> [1] "20090109"
#>
....Results when count-ing
If you set the count query, then the openFDA API will
not return full records. Instead, it will count the number of records
for each member in the openFDA field you specified for
count. For example, let’s look at drug manufacturers in the
Drugs@FDA
endpoint for "paracetamol".
count <- openFDA(search = "openfda.generic_name:ketamine",
endpoint = "drug-drugsfda",
count = "openfda.manufacturer_name.exact") |>
httr2::resp_body_json()
count$results
#> [[1]]
#> [[1]]$term
#> [1] "Civica, Inc."
#>
#> [[1]]$count
#> [1] 1
#>
#>
#> [[2]]
#> [[2]]$term
....Using count to make a time series
You can count on fields with a date to create a time series. This is
demonstrated on the openFDA website, where
they count the number of received records per day in the
drug/event endpoint. With the openFDA package, you could do
it in R like this:
resp <- openFDA(endpoint = "drug-event", count = "receiptdate")
# Extract the counts from the response as a data frame
data <- httr2::resp_body_json(resp, simplifyDataFrame = TRUE) |>
purrr::pluck("results")
data$date <- as.Date.character(data$time, format = "%Y%m%d")
# Sort on date column
data <- data[order(data$date), ]
# Restrict to data from 2000 to 2019
data <- data[data$date >= as.Date("2019-01-01") &
data$date < as.Date("2019-12-31"), ]
# Plot
plot(x = data$date, y = data$count,
xlab = "Date", ylab = "Number of adverse events reported")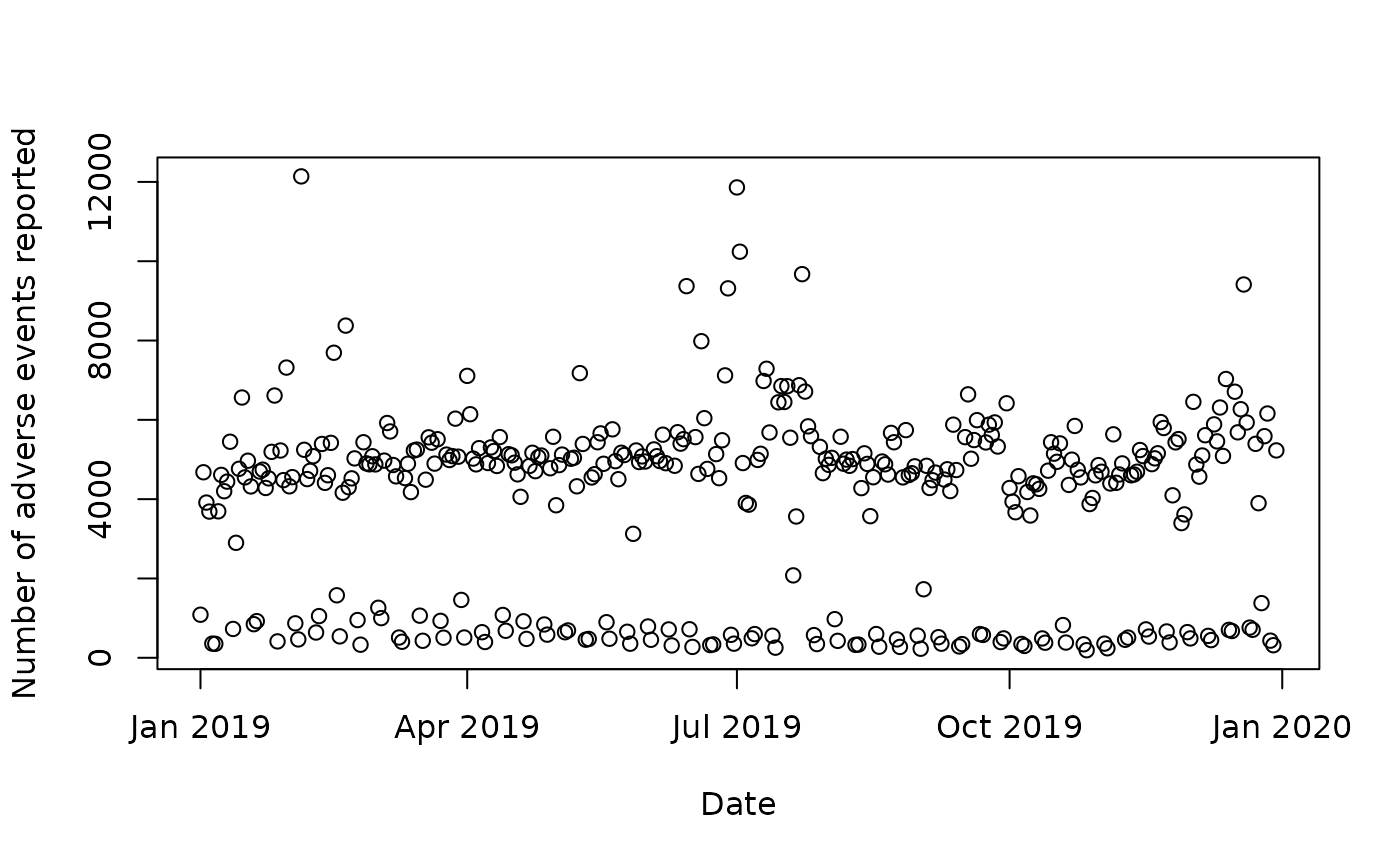
Using search terms
We can increase the complexity of our query using the
search parameter, which lets us search against specific
openFDA API fields. These fields are harmonised to different degrees in
each API, which you will need to check online.
Searching on one field
You can provide search strategies to openFDA() as single
strings. They are constructed as [FIELD_NAME]:[STRING],
where FIELD_NAME is the openFDA field you want to search
on. If your STRING contains spaces, you must surround it
with double quotes, or openFDA will search against each word in the
string. So, for example, a search for drugs with the class
"thiazide diuretic" should be formatted as
"openfda.pharm_class_epc:\"thiazide diuretic\"", or the API
will collect all drugs which have the words "thiazide" or
"diuretic" in their established pharmacological class
(EPC).
Note that there are helpers in the package
(i.e. construct_search_term()) for assembling search terms;
they are discussed below.
Let’s do an unrefined search first:
search_unrefined <- openFDA(
search = "openfda.pharm_class_epc:thiazide diuretic",
endpoint = "drug-drugsfda",
limit = 1
)
httr2::resp_body_json(search_unrefined)$meta$results$total
#> [1] 226Let’s compare this to our refined search, where we add double-quotes around the search term:
search_refined <- openFDA(
search = "openfda.pharm_class_epc:\"thiazide diuretic\"",
endpoint = "drug-drugsfda",
limit = 1
)
httr2::resp_body_json(search_refined)$meta$results$total
#> [1] 124As you can see, the unrefined search picked up 102 more results, most of which would have probably been non-thiazide diuretics.
Searching on multiple fields
The openFDA API also lets you search on various fields at once, to construct increasingly more specific queries. You can either write your own complex search queries, or let the package do (some of) the heavier lifting for you.
Write your own search term
Using the guides on the openFDA website, you can put together your own query. For example, the following query looks for up to 5 records which were submitted by Walmart and are taken orally. We can use purrr functions to extract a brand name for each record. Note that though a single record can have multiple brand names, we are choosing to only extract the first one.
search_term <- "openfda.manufacturer_name:Walmart+AND+openfda.route=oral"
search <- openFDA(search = search_term,
endpoint = "drug-drugsfda",
limit = 5)
json <- httr2::resp_body_json(search)
purrr::map(json$results, .f = \(x) {
purrr::pluck(x, "openfda", "brand_name", 1)
})
#> [[1]]
#> [1] "NAPROXEN SODIUM 220MG"
#>
#> [[2]]
#> [1] "RUGBY MUCUS RELIEF ER"
#>
#> [[3]]
#> [1] "NICOTINE MINI"
#>
#> [[4]]
#> [1] "KIRKLAND SIGNATURE ESOMEPRAZOLE MAGNESIUM"
#>
#> [[5]]
#> [1] "CHILDREN ALLERGY"Let openFDA() construct the search term
In openFDA(), you can provide a named character vector
with many field/search term pairs to the search parameter.
The function will automatically add double quotes ("")
around your search terms, if you’re providing field/value pairs like
this.
search <- openFDA(search = c("openfda.generic_name" = "amoxicillin"),
endpoint = "drug-drugsfda",
limit = 1)
paste("Number of results:", httr2::resp_body_json(search)$meta$results$total)
#> [1] "Number of results: 60"You can include as many fields as you like, as long as you only
provide each field once. By default, the terms are combined with an
OR operator in openFDA(). The below search
strategy will therefore pick up all entries in Drugs@FDA which are
taken by mouth.
search <- openFDA(search = c("openfda.generic_name" = "amoxicillin",
"openfda.route" = "oral"),
endpoint = "drug-drugsfda",
limit = 1)
paste("Number of results:", httr2::resp_body_json(search)$meta$results$total)
#> [1] "Number of results: 7052"Pre-construct a search term
You can use format_search_term() to pre-generate strings
to pass to openFDA() as search:
format_search_term(
c("openfda.generic_name" = "amoxicillin", "openfda.route" = "oral")
)
#> [1] "openfda.generic_name:%22amoxicillin%22+openfda.route:%22oral%22"Sometimes you will want to combine multiple search terms with
AND operators. To do this, set the mode
argument to "and":
search_term <- format_search_term(
c("openfda.generic_name" = "amoxicillin", "openfda.route" = "oral"),
mode = "and"
)
search_term
#> [1] "openfda.generic_name:%22amoxicillin%22+AND+openfda.route:%22oral%22"
search <- openFDA(search = search_term,
endpoint = "drug-drugsfda",
limit = 1)
paste("Number of results:", httr2::resp_body_json(search)$meta$results$total)
#> [1] "Number of results: 57"Wildcards
You can use the wildcard character "*" to match zero or
more characters. For example, we could take the prototypical ending to a
common drug class - e.g. the sartans, which are
angiotensin-II receptor blockers - and see which manufacturers are most
represented in Drugs@FDA
for this class. When using wildcards, either pre-format the string
yourself without double-quotes or use
format_search_term() with exact = FALSE. If
you try to search with both double-quotes and the wildcard character,
you will get a 404 error from openFDA. In the code block below, we use a
wildcard to find all drugs with generic names ending with
"sartan", and count on the manufacturer names.
search_term <- format_search_term(c("openfda.generic_name" = "*sartan"),
exact = FALSE)
search <- openFDA(search = search_term,
count = "openfda.manufacturer_name.exact",
endpoint = "drug-drugsfda",
paging = "always")
results <- httr2::resp_body_json(search, simplifyDataFrame = TRUE) |>
purrr::pluck("results")
head(results, n = 10)
#> term count
#> 1 Alembic Pharmaceuticals Limited 15
#> 2 Macleods Pharmaceuticals Limited 15
#> 3 Alembic Pharmaceuticals Inc. 14
#> 4 Aurobindo Pharma Limited 13
#> 5 Lupin Pharmaceuticals, Inc. 10
#> 6 Mylan Pharmaceuticals Inc. 7
#> 7 Solco Healthcare US, LLC 7
#> 8 Zydus Lifesciences Limited 7
#> 9 Ascend Laboratories, LLC 6
#> 10 Camber Pharmaceuticals, Inc. 6It looks like "Alembic Pharmaceuticals" is very active
in this space - interesting!
Large result sets
The openFDA API only returns up to 1000 results at one time. To
return a results set larger than 1000 records, a basic paging system is
implemented. On a per-call basis, this is controlled by the
paging parameter in openFDA(). To change this
behaviour for the duration of an R session, edit the option
"openFDA.paging", as demonstrated above.
This argument must take one of these values:
-
"ask"(default; only valid if R is running interactively) - ask before performing paging -
"yes"- always perform paging where necessary, but print a message to the console -
"yes-quiet"- always perform paging where necessary, but don’t print a message to the console -
"no"- never perform paging, but print a message to the console -
"no-quiet"- never perform paging, but don’t print a message to the console
When paging is performed, openFDA() returns a list of
httr2 response objects, instead of a single response:
resps <- openFDA(search = "openfda.manufacturer_name:Pharma",
limit = 750,
endpoint = "drug-drugsfda",
paging = "always",
paging_verbosity = "verbose")
#> ! openFDA returned more results (1991) than your `limit` (750).
#> ℹ With 750 results per page, pagination will require 3 queries to retrieve all
#> of these results. This will take at least 1s.
resps
#> [[1]]
#> <httr2_response>
#> GET https://api.fda.gov/drug/drugsfda.json?search=openfda.manufacturer_name:Pharma&limit=750
#> Status: 200 OK
#> Content-Type: application/json
#> Body: In memory (3510953 bytes)
#>
#> [[2]]
#> <httr2_response>
#> GET https://api.fda.gov/drug/drugsfda.json?search=openfda.manufacturer_name%3APharma&limit=750&skip=0&search_after=0%3DNEwDMJgBCsORzztADuLN
#> Status: 200 OK
#> Content-Type: application/json
#> Body: In memory (3678214 bytes)
#>
#> [[3]]
#> <httr2_response>
#> GET https://api.fda.gov/drug/drugsfda.json?search=openfda.manufacturer_name%3APharma&limit=750&skip=0&search_after=0%3Dk0wDMJgBCsORzztAFfnR
#> Status: 200 OK
#> Content-Type: application/json
#> Body: In memory (2440677 bytes)Other openFDA API features
This short guide does not cover all aspects of openFDA. It is recommended that you go to the openFDA API website and check out the resources there to see information on: